Genome Evolution of Structural Variants
Structural variants (SVs) are large (>50 bp) mutations, e.g., deletions, insertions, and inversions, that more likely change phenotypes than single nucleotide variants (SNVs) and, thus, are subject to natural selection and important in evolution. I use long-read sequencing to resolve complex SVs, apply phylogenetic and population genetic methods to infer their evolutionary histories, and design statistical models to determine their functional significance in organisms.
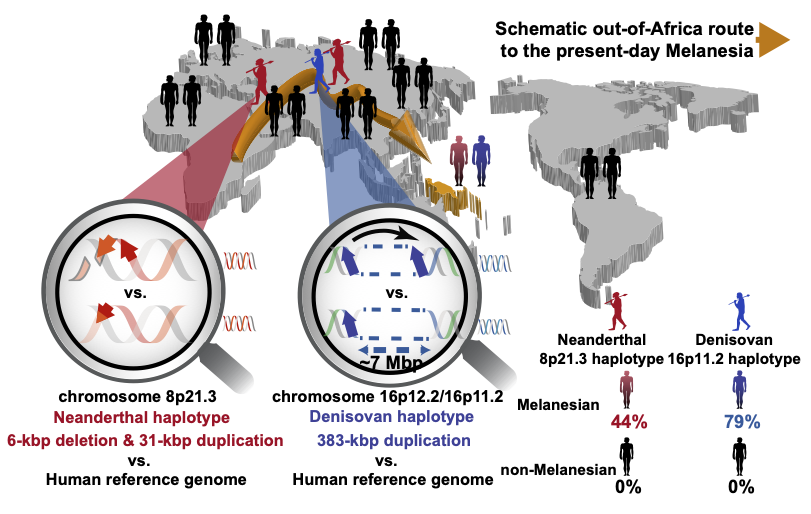
Adaptive SV Introgression in Humans
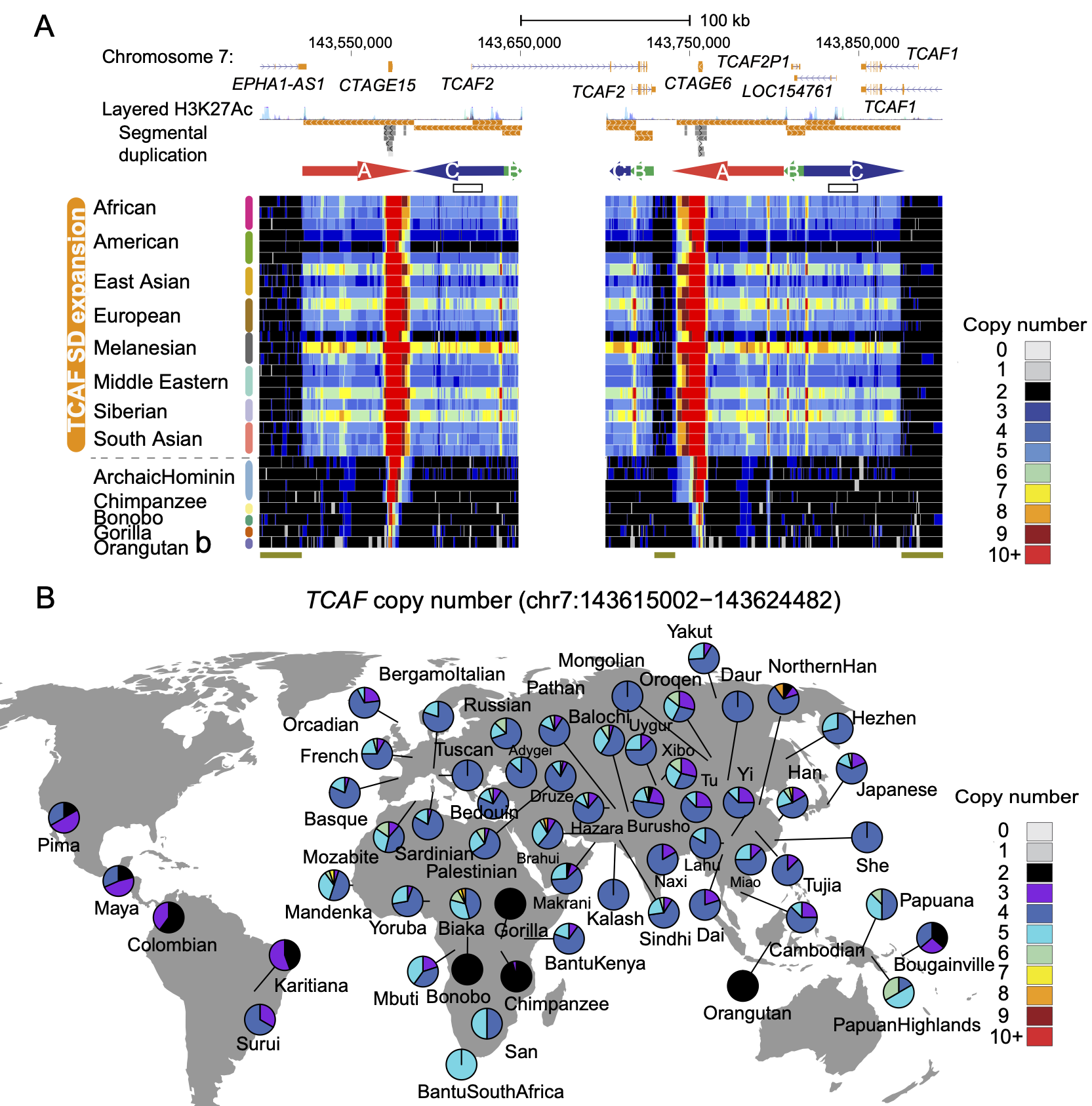
Human-specific Expansion of Thermoregulatory Genes
Recurrnt Inversions in Great Apes
Learning Evolution using Statistical Modeling
DNA variation data record useful information about the evolution of organisms. I design and apply quantitative and statistical methods to reconstruct the evolutionary history in human and nonhuman primates as well as other organisms. Understanding evolution is a critical step towards understanding the biological world that we live in and helps us to understand the past and predict the future.
Human Origin in Africa
Human-specific Expansion of Thermoregulatory Genes
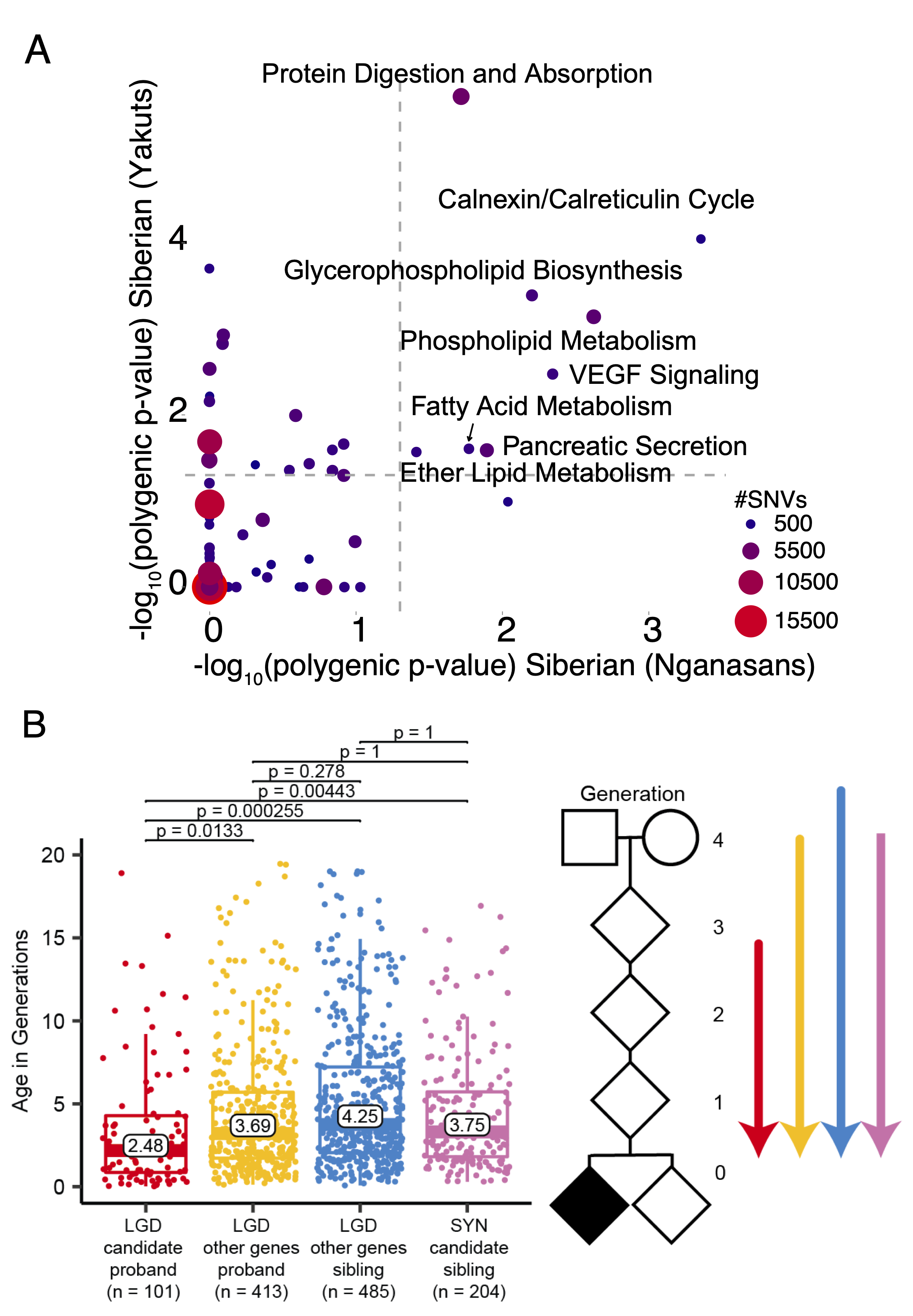
Human Evolution in Cold Environments
Evolution and Speciation of Desert Tortoise
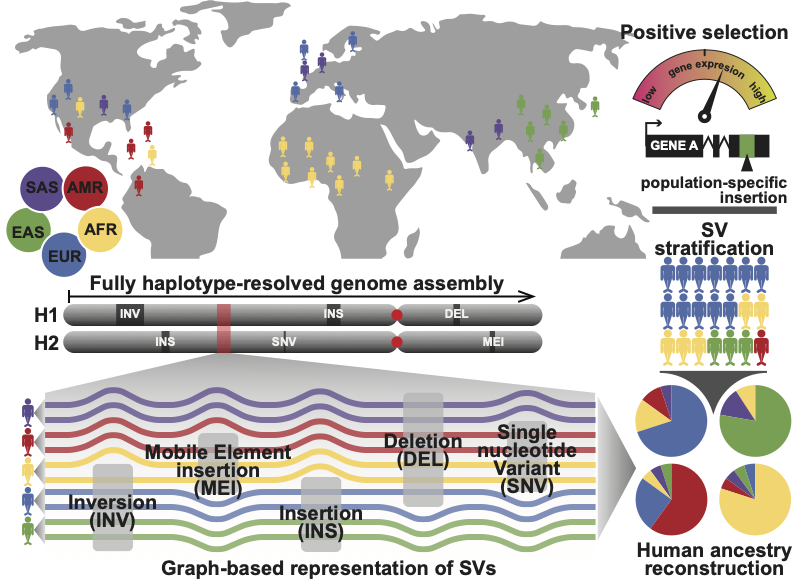
Delineating Genome Diversity and Evolution using Long-Read Sequencing
20 years after the first draft of human genome, with the recent development of long-read sequencing, we now finally have the ability to assemble diploid genomes and study complex genomic regions for the first time. As part of efforts from the Human Genome Structural Variation and Human Pangenome Reference Consortia, my research program leverages these resources to better capture the genetic diversity of our species, especially in regions of more complex forms of variation that short-read data cannot ascertain.
Adaptive SV Introgression in Humans
Haplotype-resolved Diverse Human Genomes
Complete Linear Assembly of Human Autosome
Evolutionary Medicine
Evolution is critical for understanding human health, from how populations adapt to different environmental niches and against pathogens to the genetic predisposition to diseases. Using population genetics methods, we identified genetic variants that may affect bone and muscle synthesis in African pygmies as well as those involved in fat metabolism that could contribute to the cold adaptation in Siberian hunter–gatherers. In a large cohort of autism families, we demonstrated that ultra-rare likely-gene disruptive (LGD) variants in probands are significantly younger than those same type of variants in siblings and that many such variants are under strong purifying selection and act on a distinct set of genes not yet associated with autism.
Evolutionary Medicine - Autism
Human Evolution in Cold Environments
Adaptations in African Pygmies